The BIDS EEG specification follows the general BIDS data structure originating from MRI with some modifications to accommodate EEG data types.
File formats
- BIDS encourages “official” use of The European Data Format (EDF) and Brain Vision Core Data Format files for the electrophysiological recording data.
- An “unofficial” option is to use MATLAB EEGLAB toolbox files (.set and .ftd) or the Biosemi format (.bdf).
- JSON sidecar files contain dictionary-type metadata.·
- Tabular data is stored using tab separated files (.tsv).
Each subject has a directory of raw data containing subdirectories for each session and modality. At the root level of the directory, the README, CHANGES, and dataset_description.json files provide basic information about the dataset, and a metadata file with the suffix _eeg.json, that specifies the task, the EEG system used (amplifier, hardware filter, cap, placement scheme, etc.).
Any metadata (.json, .bvec, .tsv, etc.) may be defined at any directory level. The values from the top level are inherited by all lower levels unless they are overridden by a file at the lower level. A participants.tsv data file is accompanied by a participants.json file, which contains the description of the columns in its associated .tsv file. Usually, each .tsv file is accompanied by a .json file that provides metadata.

A stimuli directory contains the stimuli that were presented to the participants in the experiment.
Example:
- Stimuli
- Stim1.png
- Stim2.png
If BIDS does not support the original data, it can be included in an additional sourcedata directory. A sourcedata directory containing the original behavioral and EEG data can be present, as well as a stimuli directory and a code directory to allow data conversion and preprocessing to be reproduced.
The EEG data are saved per subject (e.g., Sub-01 directory) within the eeg subdirectory. The eeg subdirectory contains the EEG and metadata. For instance, for a single session study, Sub-XX would have subdirectory eeg which contains multiple EEG files as Sub-XX_task-YY_run-ZZ_eeg.edf files corresponding the different runs of EEG data acquisition. In addition, Sub-XX_task-YY_channels.tsv must be specified describing the parameters of the data acquisition. Sub-XX_taskYY_electrodes.tsv and Sub-XX_task-YY_coordsystem.json files should be specified, if the positions of the electrodes are known.
Example:
- SUB-01
- EEG
- Sub-01 task-TASKNAME eeg.edf
- Sub-01 task-TASKNAME eeg.json
- Sub-01 task-TASKNAME events.tsv
- Sub-01 task-TASKNAME channels.tsv
- Sub-01 task-TASKNAME electrodes.tsv
- Sub-01 task-TASKNAME coordsystem.json
Electrodes versus Channels
The terms electrodes and channels are sometimes used interchangebly by EEG researchers. It is important when using the BIDS-EEG structure to differentiate between the two terms to ensure for clarity. The term electrode refers to the physical component that picks up the electrical signal on the scalp. Channel refers to the signal that is recorded by an electrode. Designate channels with a channels.tsv. file and electrodes with an electrodes.tsv file.
Fiducials, Anatomical Landmarks, Coordinate System
In addition to the blended synonymous usage of the terms ‘electrodes’ and ‘channels’ among EEG-researchers, fiducials and anatomical landmarks are often interchanged in a similar way. Researchers use fiducials to aid electrode localization, as well as co-registration. In co-registration, other types of data such as the participants T1 structural MR scan, a T1 template image, or other types of models are used.
The term anatomical landmarks refers to characteristic locations that one can locate with the human eye on the research participant, such as the nasion (where the two nasal bones meet the frontal bone).
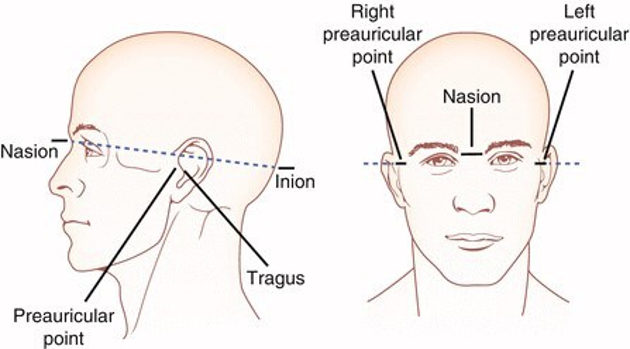
A coordsystem.json designates fiducials, anatomical landmark location, the coordinate system, and units that the coordinate system and landmarks uses.
The coordsystem.json is required when the (optional) electrodes.tsv is used. If the T1 anatomical MR scan is available, then the position of landmarks and fudicials as they relate to the image are indicated in the T1w.json file included with the MRI data.
Conversion Tools for EEG:
Several tools have been developed to convert data to the desired EDF (16 bit) or BrainVision data format (32bit). MATLAB’s FieldTrip Toolbox has a data convertor utility that converts a variety of EEG formats to the EDF and BrainVision data format.
Those who prefer Python can use the following tools for for conversion to EDF and BrainVision data formats.
EEGLAB for MATLAB, is an open source tool for EEG analysis. The function std_tobids.m, converts an EEGLAB study, commonly used for group analysis, to the BIDS-EEG specification. Sub-functions automate the creation of channel and electrode .tsv and .json files are also available.
Most EEG formats can be used in FieldTrip, and this includes those converted to the BIDS standard. It can also convert files to the EDF and BrainVision formats. A benefit to the analysis of the data output in BIDS standard is the availability of useful metadata (events and annotations), readable from the events.tsv sidecar file. FieldTrip also has a data2bids.m function that aids in conversion of EEG, MEG, iEEG and MRI data to BIDS and ensures proper metadata is provided. Tutorial for BIDS using FieldTrip can be found here.
MNE is an open access, Python based tool for visualization and analysis of EEG data with functions that read the BIDS EEG data formats. A near-complete convertor to BIDS is available that assists in getting the raw data into the basic BIDS format with the necessary metadata files.
SPM12 includes a library, spm_BIDS.m, that can be used with BIDS datasets.
