Raw iEEG data types and structure
There are two raw data formats that are output by the acquisition systems most commonly used for iEEG by researchers at UiO. The following section provides a brief overview of their content and purpose.
Neuralynx uses an acquisition software called Cheetah. Neuralynx system outputs its own proprietary file formats. Its standard setting is to create a separate file for each individual channel. These channel files are written into a ‘dataset directory’ and the files therein are able to be read concurrently. The Fieldtrip toolbox contains reading functions that work with these file types. See http://www.fieldtriptoolbox.org/getting_started/neuralynx/ for a listing of the exact functions.
Neuralynx file types

Micromed also has its own acquisition and analysis software, Brain Quick EEG. The system permits two types of raw data formats as outputs.
Micromed file types

Localization: MRI, CT and Photographic Raw Data
Structural magnetic resonance imaging (MRI) scans are often made of participants to aid localization. Some studies may also combine scalp EEG, iEEG and fMRI. For example, fMRI may be used in advance of electrode placement to identify networks and specific areas of interest (Fox et al., 2018). Thus, it is important to also have some basic understanding about MRI data as well. For a more thorough introduction to (f)MRI data, please see our guide on the topic.
Without knowing the precise placement of the electrodes, claims to the superior spatial resolution of iEEG are difficult to make (Lachaux et al., 2003). For iEEG, structural MRI scans are often made after surgical implant of the electrodes to assist in electrode localization, i.e. the labeling of anatomical placement as it pertains to the individual, strip or mesh of electrodes. A cortical mesh can be constructed by using the normalized structural scan and co-registering EEG data according to the fiducials (Henson et al., 2019). Because of the very individual nature of iEEG electrode placement, researchers will likely have to plot out the location of electrodes manually with the assistance of a graphic interface. Photographic images taken during surgery, both before and after electrode placement can also greatly assist in this task (Dalal et al., 2008). It is important that the MRI images be taken after surgical implantation if possible, because of the brain-shift phenomena, which results from the surgical procedures, electrodes themselves and post-operative swelling. This can cause electrode displacement. This problem is most likely to occur with grid and strip electrodes and is less common in depth electrodes. T1 MRI scans taken post-implantation are the best method to use in terms of localizing electrodes, as brain-shift is less of a problem. However, in some cases electrodes may be difficult to identify in a T1 scan because they can appear dark and illdefined due to artifacts caused by the electrodes themselves. In such cases, coregistration to a post-operative CT and pre-operative MRI scan may be preferred (Lachaux et al., 2003; Stolk et al., 2018).
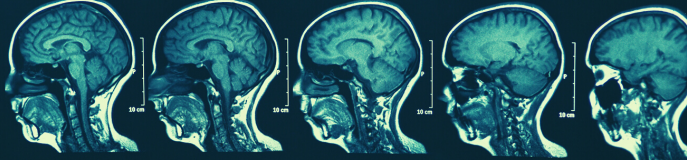
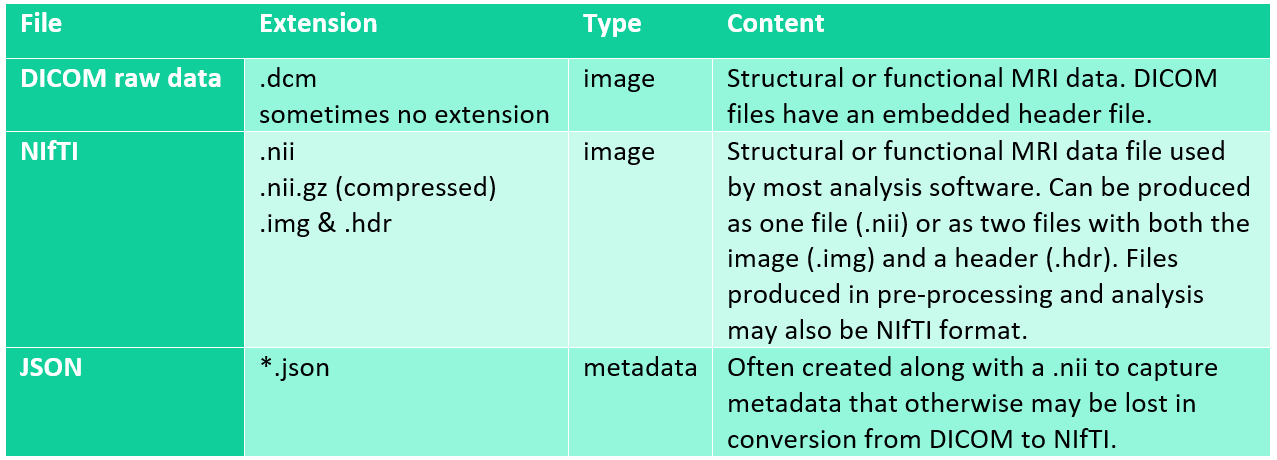
The raw data which is taken directly from the scanner is typically extracted as DICOM files. DICOMS entail a very high volume of data. Thus, one should ensure that the password-protected, encrypted hard drive used for transferring files from the scanner to Lagringshotell is large enough to account for this. DICOMS must be converted to NIfTI files, which is the format used by most modern neuroimaging analysis software. DICOMS may need to be sorted and re-named prior to conversion to NIfTI. Some conversion applications will automatically create a single NIfTI file from the DICOM images (.nii), while others may offer a two file option with an image file (.img) and a header (.hdr). Alternately, JSON files can be created by some programs to supply metadata that may be lost in DICOM conversion.
Data Extraction and Transfer
DICOM and iEEG data are temporarily stored on a password-protected, encrypted hard drive at the point of collection. It is important to retrieve this data on the day of data collection whenever possible. For example, DICOM images are routinely removed from the scanner console and will no longer be available for transfer unless arrangements are made with the radiographers in advance. The encrypted hard drive will then typically be delivered to the data manager. The data manager will then anonymize the data by removing any remaining sensitive information from header files and defacing the MRI scans. The original images will be kept in TSD and the data will be deposited in the durable folder on Lagringshotell for use. It is important that you make a copy of this data and do not work with data stored in the durable folder directly. You may instead copy the data to your own user folder on Lagringshotell for preprocessing and analysis. The data should never be stored on personal computers, and must remain in Lagringshotell.